The Proteomics and Metabolomics Program
- 19057
- (202) 25351500, Ext. 7226
- [email protected]
FAQ
Why should I consider proteomics in my research?
Proteomics is a branch of the “Omics” high throughput sciences that is growing rapidly. The future of Omics technologies started already to overshadow the traditional analytical methods. No doubt, adding a proteomics in your research will boost your output, scientific impact and increase possibility to publish in a high impact factor journals.
Where is the Proteomics unit?
We are located on the second floor of the Children’s Cancer Hospital Egypt 57357. A well-known landmark in Egypt.
How do I use the Proteomics Laboratory?
Start by e-mailing us to discuss your project. Consultation is free. We will help you identify experimental designs most likely to meet your objectives and most compatible with our methods and instrumentation.
Collaboration or service?
We charge our service. Exceptionally, we may consider collaboration with partial or complete waiving of the costs based on the potential interest we have as 57357 in the work.
What is the cost of the service?
After consultation meeting, we will recommend you the optimal working platform to work with. This will include also the costing of the sample.
How I can get the result?
When analysis is done, we will inform you. A report will be released with a softcopy of the data itself. We don’t deliver the data through internet.
Does the proteomics laboratory do service analysis for non-CCHE-57357 clients?
Yes, one of our duties as an NGO organization is to help researchers to conduct their research efficiently with high standards.
What is the flow of work?
Once you asked for consultation and agreed on the work protocol, we will ask you to send the samples after instructing you on preparation method. Once we received the samples, we will start assigning an expert to process and analyze your samples. Following this step, a report is generated and revised by the unit head, upon approval, the report with data is delivered to the researcher.
Can I attend processing and analysis steps?
No, we don’t allow attending sample processing or analysis. This is simply because it might increase error and loss of concentration. Exceptional approval might be considered if necessarily.
I can’t understand the results, what can I do?
We do support you to understand the result. Face to face consultation is by appointment, alternatively, communicate by email and we will reply within 48 hrs.
How could I represent my data?
We help you fully understand your result and present it. Additionally, offer an extra service for data presentation. A unique platform called R. Using R language allows you to boost your result presentation as seen in well-known scientific journals such as Cell, Science and Nature. We advise researchers to see the power of R language by googling “R graphics”
How we assure quality control up to international proteomics guideline standards?
CCHE-57357 guarantee the highest quality control standards. Here in the proteomics unit, we calibrate the mass spectrometry prior to any analysis, run blanks to make sure that our mass spec is clean and free from any turn overs, apply blanks between runs and apply an automatic calibration between samples when processing time is long. Following completion of the analysis, each chromatography is inspected manually for some detailed parameters to make sure samples are analyzed perfectly. Raw files are securely transferred to our bioinformatics tools for protein assembly. Generated data is reported and revised by two researchers then approved by the head of the unit.
How can I be sure about bringing a correctly processed metabolite extract for service analysis?
Discuss your sample extraction protocols with lab members. Please follow the international protocol depending on your samples. We can direct you to the appropriate methodology via a free- of –cost consultation.
How many metabolites can I measure in my sample?
This depends on the kind of biological sample and the followed metabolite extraction protocol. The list of detectable metabolites can be found in the Mass bank spectral library. We have been working in the last few month to construct our own curated database. A fruitful sold and powerful metabolite database is now ready to enrich your results.
Can I identify metabolites of my interest?
Yes, this is possible in a non-targeted workflow but it requires additional time, data analysis tools and LC-MS runs. Please discuss this with lab members in advance.
Why the analysis did not identify many metabolites in my sample?
There are several reasons depending on the type of biological sample. If your sample is enriched with specific peaks compared to a control or blank sample, but no metabolite has been identified, then it’s chiefly the limitation of database available. We can perform more elaborated studies using an open source database as required. Furthermore, the use of database search or exact mass filtering (with HMDB, LIPID MAPS, Mass bank, Metfrag, etc.) may help identify features for which fragmentation spectra has not been acquired. The database search will provide an idea about the possible metabolite identity. As a confirmatory step, we may acquire and match spectra from the corresponding commercially available standards.
Why no metabolites have been detected in my sample?
The reasons could be either dilution of samples, loss of metabolites during sample preparation (i.e. metabolite extraction procedure) or solubility issues during reconstitution of dried sample. Please make sure that your sample amount meets the requirements and protocol of verification accepted before proceeding with your precious samples. It is highly recommended to discuss your sample extraction protocols at the early phase with the facility staff. Use high grade solvents to minimize the background noise.
How reliable is the provided information of the identified metabolites?
We are performing all the identifications based on in-house generated metabolite spectral libraries with mass high accuracy and fragmentation pattern (MS/MS) matching. Mass spectrometry has its inherent limitation in identifying structural and chiral isomers unless well separated by chromatography.
What kind of results will be provided from metabolomics analysis for my sample?
You will receive a report in Excel format with a list of identified metabolites, mass over charge (m/z) values, chromatographic retention times (RT) and peak areas / intensities. In an untargeted workflow, you will be provided with a list of all detected features m/z, RT and peak areas / intensities) both with and without metabolite identifications. If you have an experimental design with specific sample groups then we can also provide you fold change along with p and q values. The acquisition method, raw data files, chromatograms and mass spectra will be also provided.
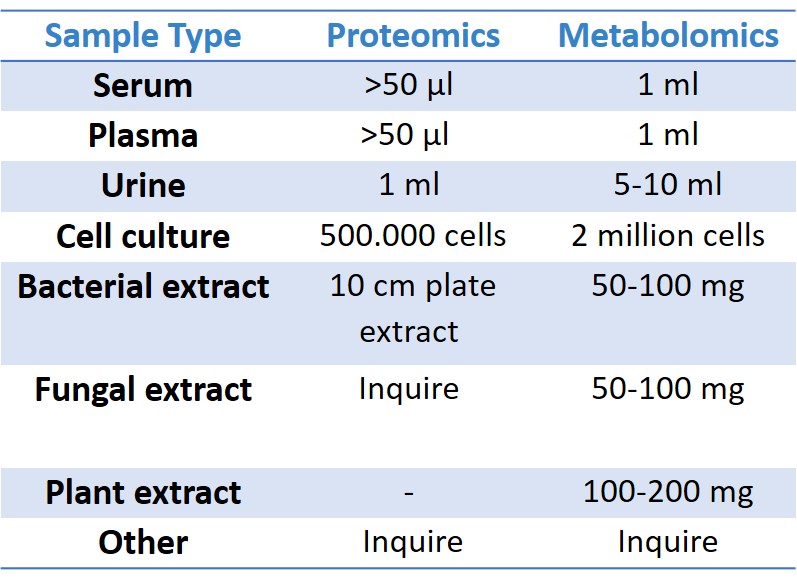
What is the minimum amount of protein required for MS analysis?
This is the single most frequent question we get and the hardest one to answer. The correct boring answer is “it depends on the nature of the sample”. Although we can detect as low as femtomole level, we recommend to send a “gram” scale protein extract. For further details, kindly check the below table.
- Research Program Overview
- Research Interest
- Research Pipelines
- Instruments and Software
- Photo Gallery
- Video Gallery
- Services and Costs
- Grants
- Workshops
- Publications
- Meet Our Team
- Partnership
- Conferences
- Alumni
- Lab Protocols
- Review
- Do's and Don'ts
- FAQ
- Careers
- In-House Bioinformatics Tools
- For Inquiries