The Proteomics and Metabolomics Program
- 19057
- (202) 25351500, Ext. 7226
- [email protected]
Services and Costs
Services and Costs
We offer a unique service with different assay length and applications. Our uniqueness stems from the fact that we are simply the only ones offering these services in Egypt. Costs include reagents, sample processing, manpower, instrument usage, data analysis and reporting.
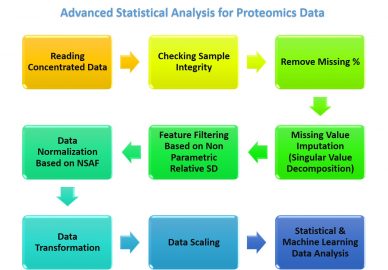
General workflow
We work professionally according to world standards to ensure accurate, sensitive results. For all samples we perform an efficient protein extraction using varieties of extraction buffers, quantification of proteins and appropriate protein reduction and alkylation prior to trypsinization. All samples are subjected to C18 desalting step to make sure purified peptides are going to our clean TripleTOF 5600+.
Data analysis
- Data is searched by up to two tandem mass spectrometry protein identification algorithms, including X!Tandem, OMSSA, comet, tide, andromeda.
- Label free quantification based on spectral abundance “NSAF or emPAI” or area under the curve “AUC” is provided free of charge
- Samples or groups of samples are compared, generating fold-change values for every measured protein.
Service 101 : (consultation)
Free (requires appointment)
Service 102: (Proteome profiling, 20-30 min)
- Assay: (365 Proteome profiling)
- Specifications: nanoLC-MS/MS, 10,000 sequencing events, 15 cm column, 20-30 min gradient.
- Sample types: Affinity-purified samples or characterization of individual proteins
Service 103: (Proteome profiling, 60 min)
- Assay: (365 Proteome profiling)
- Specifications: nanoLC-MS/MS, 50,000 sequencing events, 15 cm column, 60 min gradient.
- Sample types: sub-cellular fractions, exosomes, prokaryotic organisms
Service 104 : (Proteome profiling, 90 min)
- Assay: (365 Proteome profiling)
- Specifications: nanoLC-MS/MS, 150,000 sequencing events, 15 cm column, 90 min gradient.
Service 105 : (Proteome profiling, 180 min)
- Assay: (365 Proteome profiling)
- Specifications: nanoLC-MS/MS, 250,000 sequencing events, 15 cm column, 180 min gradient.
- Sample types: Eukaryotic organisms, biofluid plasma/serum/urine/saliva profiling, host-pathogen profiling and microbiome profiling, and other samples consisting of a mixture of organisms.
Service 106 : (Library building)
- Assay: (365 Proteome profiling- Library building)
- Specifications: nanoLC-MS/MS, 450,000 sequencing events, 15 cm column, 180 min gradient in a replicates (3-5) until saturation.
- Sample types: Ideal for building a comprehensive proteomic library of a single organism for maximum sequence coverage and post-translational modification characterization
Service 107 : (Lane/band profiling)
- Assay: (“Per- band/ Per lane” sequencing)
- Specifications: nanoLC-MS/MS, 15 cm column, 30 min gradient per slice/band.
- Sample types: Ideal for one dimensional and 2 dimensional electrophoresis
Service 108 : (PTM)
- Assay: (Post-translational modification (PTM) analysis)
- Specifications: nanoLC-MS/MS, 15 cm column, 60-120 min gradient depending on sample complexity.
- Sample types: Ideal for one dimensional and 2 dimensional electrophoresis
Service 109 : (IP/MS analysis)
- Assay: (IP/MS analysis)
- Specifications: nanoLC-MS/MS, 15 cm column, 60-120 min gradient depending on sample complexity.
- Sample types: Antibody based capturing coupled with MS analysis
Service 110 : (Relative quantification analysis)
- Assay: (relative quantification analysis)
- Specifications: nanoLC-MS/MS, 15 cm column, 60-120 min gradient depending on sample complexity.
- Sample types: SILAC for cell culturing and dimethyl- labeling works will most biological samples.
Service 111 : (MRM/SRM analysis)
- Assay: (MRM/SRM analysis)
- Specifications: nanoLC-MS/MS, 15 cm column, pilot exploratory run followed by targeted 30- 60 min gradient.
- Sample types: any biological sample.
Service 112 : (Plasma profiling)
- Assay: (Plasma profiling)
- Description: DDA (Data dependent acquisition method). The sample is depleted from highly abundant proteins such as albumin and globulin then subjected to Lc-MS/MS analysis.
- Specifications: nanoLC-MS/MS, 15 cm column, 60-180 min gradient depending on sample complexity.
- Sample types: plasma/ serum
Service 113 : (SWATH technology)
- Assay: (SWATH technology)
- Description: DIA (Data independent acquisition method) that captures most peaks in the sample and searched against a library.
- Specifications: nanoLC-MS/MS, 15 cm column, 60-180 min gradient depending on sample complexity.
- Sample types: any biological sample/fluid
Service 114 : (Pharmaceutical and small molecule analysis)
- Assay: (Pharmaceutical and small molecule analysis)
- Description: TDM, drug delivery, pharmacokinetics and other.
- Specifications: Customized depending on experimental.
- Sample types: any biological sample/tablets and other drug formats
The service provides three types of analysis:
- Positive mode.
- Negative mode.
- Both modes; with a 30% reduction in the service cost.
Service 115 : (Adulteration, Forensic, Pesticide)
- Assay: (Adulteration, Forensic, Pesticide)
- Description: testing adulteration in juice, detecting pesticide levels in food and checking drug residues in urine.
- Specifications: Customized depending on experimental.
- Sample types: any biological sample/tablets and other drug formats
Service 116 : (In-depth analysis)
- Assay: (Comprehensive report “In-depth analysis”)
- Specification: For Proteomics or metabolomics analysis, a set of comprehensive tests and software “see instruments and software tab” will be used to generate a ready publishable data and graphics. This utilize a set of bioinformatics data analysis software and R language. More details are available upon request.
Show Cases

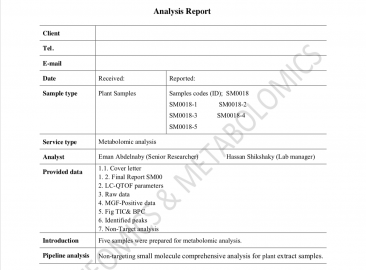
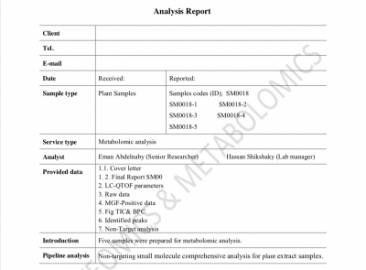
Model Report 
Model report (Proteomics) 
Gene Ontology and enrichment analysis 
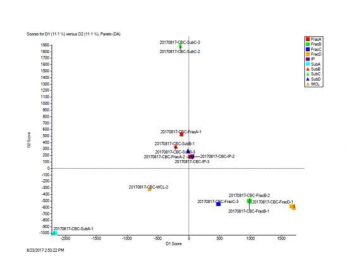
Marker View 
Peak View 
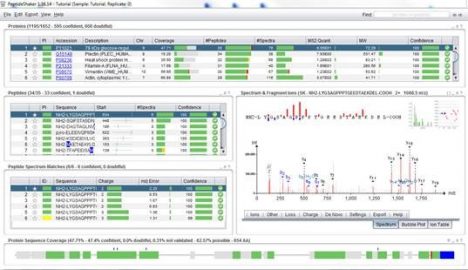
Peptide Shaker 
Peptide Shaker 
Peptide Shaker 
Protein pilot 
Protein-pilot 
Quality control analysis 
Statistic analysis
Service Request Form: Step 1
Service Request Form: Step 2
To be filled only after you have received an email from us that we have accepted your sample.
Click here to download in the form
Consent form application tutorial:
Service Tracking System
- Research Program Overview
- Research Interest
- Research Pipelines
- Instruments and Software
- Photo Gallery
- Video Gallery
- Services and Costs
- Grants
- Workshops
- Publications
- Meet Our Team
- Partnership
- Conferences
- Alumni
- Lab Protocols
- Review
- Do's and Don'ts
- FAQ
- Careers
- In-House Bioinformatics Tools
- For Inquiries